Software
Escort: Data-driven selection of analysis decisions in scRNA-seq trajectory inference
Evaluates a single-cell RNA-seq dataset’s suitability for trajectory inference and quantifies trajectory properties influenced by analysis decisions.
scLANE: Interpretable trajectory inference testing
Identifies differential expression of genes over a trajectory and characterizes dynamics using interpretable model coefficients. scLANE can be implemented downstream of any pseudotime or RNA velocity method.
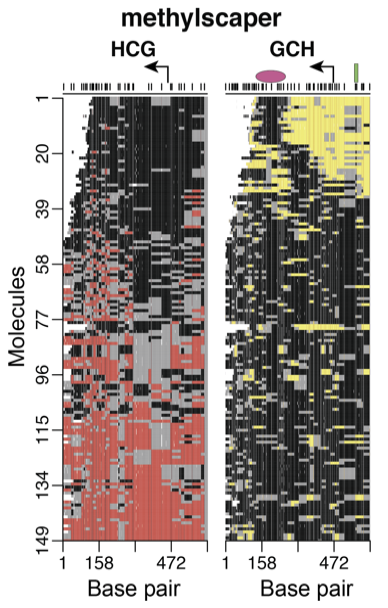
methylscaper: joint visualization of DNA methylation and nucleosome occupancy in single-molecule and single-cell data
Optimally orders molecules/cells in an interactive visualization application to discover methylation patterns, nucleosome positioning, and transcription factor binding sites.
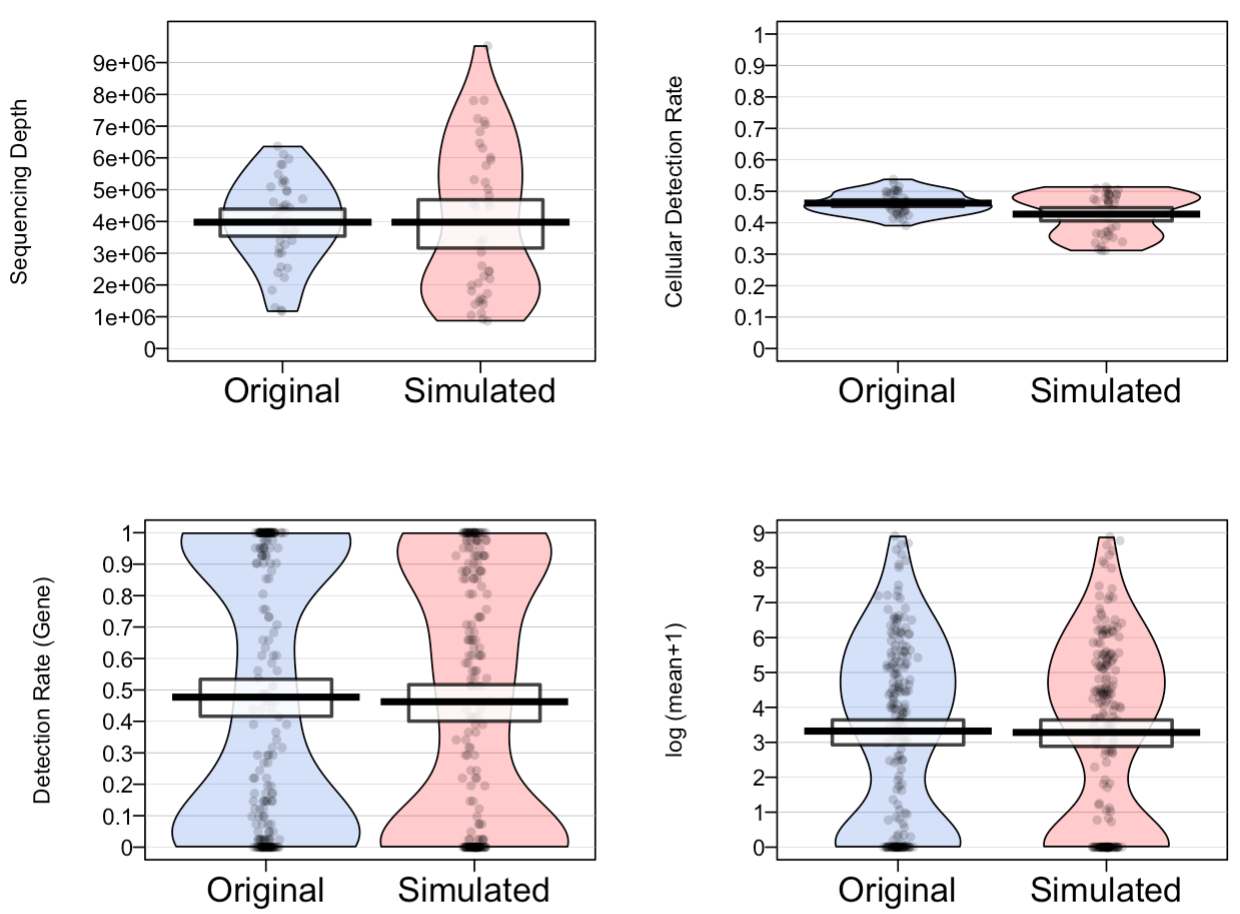
Scaffold: data-generation based simulation of single-cell RNA-seq data
Generates scRNA-seq data by statistically modeling the experimental data generation process. Scaffold simulates non-UMI, UMI, and 10X data.
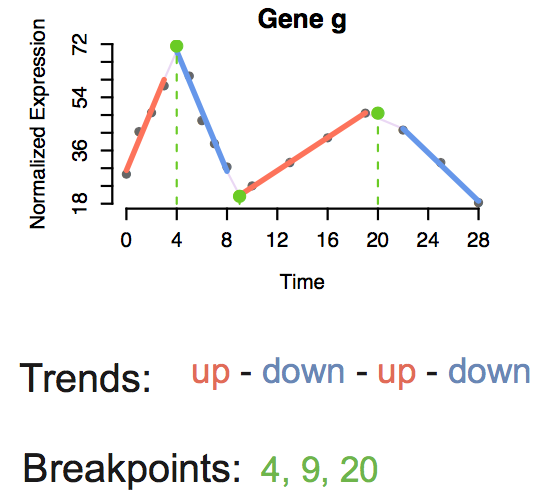
Trendy: Analysis of expression dynamics for high-throughput ordered profiling experiments
Characterizes each gene’s expression pattern and summarizes overall dynamic activity in time-course experiments (RNA-seq, microarray, etc.). An interactive application allows visualization of individual gene’s dynamic trends.
SCnorm: normalization for single-cell RNA-seq data
Normalizes scRNA-seq data by estimating group-specific scaling factors where gene groups are defined based on their estimated count-depth relationship.